Passive Device Simulation
1. How to create a new SDK simulation project?
First, we need to import the SDK package to perform optical simulation calculations.
import maxoptics_sdk.all as mo
Following that, as shown in the following code, create a simulation project. The name represents the project's name.
# region --- Project ---
pj = mo.Project(name=project_name)
# endregion
2. How to add materials in SDK simulation?
Next, we need to add the materials used in the simulation process, as shown in the following code. Here, name represents the material's name, data represents the material object from the material library, and order is the priority of material coverage. When structures of different materials overlap, materials with higher order values will take precedence over those with lower values. In the case of equal values, the one added later takes precedence.
# region --- Material ---
mt = pj.Material()
mt.add_lib(name='Si', data=mo.Material.Si_Palik, order=2)
# endregion
2.1 How to add a non-dispersive material?
Additionally, we can add a non-dispersive material using add_nondispersion. The data in data=[(real, imag)] represents the real and imaginary parts of the refractive index.
mt = pj.Material()
mt.add_nondispersion(name="SiO2", data=[(1.444, 0)], order=1)
2.2 How to add a dispersive material?
We can add a dispersive material using add_dispersion, as shown in the following code. data=[(wavelength, real, imag)] represents the real and imaginary parts of the refractive index at different wavelengths. we can perform fitting to obtain the refractive index of the material at different wavelengths.
mt = pj.Material()
mt.add_dispersion(name="SiO2",data=[(1.55e-06, 1.444, 0), (1.30e-06, 1.81, 0.227)], order=1)
2.3 How to add an anisotropic material?
We can add an anisotropic material using add_anisotropy. In this case, data=[(wavelength, nx_real, nx_imag, ny_real, ny_imag, nz_real, nz_imag)] represents the anisotropic refractive index's real and imaginary parts at different wavelengths. Similarly, we can use fitting to obtain the anisotropic refractive index of the material at various wavelengths.
mt = pj.Material()
mt.add_anisotropy(name="LN", fitting=None,
data=[(1.55e-06, 2.211, 0, 2.138, 0, 2.211, 0)], order=2)
3. How to add geometric structures in SDK simulation?
Next, let's learn how to create a geometric structure in SDK.
3.1 How to add background refractive index?
Define the background refractive index in Structure as shown below.
# region --- 3. Structure ---
st = pj.Structure()
# endregion
3.2 How to add a rectangular structure?
As shown in the code below, we can add a structure within the Structure function using add_geometry. To add a rectangular structure, use type='Rectangle'. Specify the material from the previously defined material types, and set the mesh_order to determine the material coverage priority, as explained earlier in the material addition section. Within the parameters of the geometry section, x/y/z represent the center coordinates of the structure in three directions, while x_span/y_span/z_span determine the width of the structure in these three directions.The tilt_position is to ensure that the models are placed at the specified sizes on the different ratio of sidewalls when building the structures. Selections are ['top', 'bottom', 'middle','user_defined'].The tilt_angle is tilt angle of structure sidewall.Under the selectionuser_defined, we also need to set the ratio by user_defined_position.
st.add_geometry(name="rectangle", type="Rectangle",
property={
"material": {"material": mt["Si"], "mesh_order": 2},
"geometry": {"x": 1, "x_span": size,"y": space, "y_span": wg_width, "z": 1, "z_span": wg_height,
"tilt_position":"user_defined",
"tilt_angle":70,
"user_defined_position":0.1,}})
3.3 How to add an arc waveguide in SDK simulation?
Similarly, we can add an arc waveguide structure in SDK. In the geometry section, we usetype="Ring" to build an arc waveguide.The inner_radius and out_radius represent the inner and outer radius of the arc structure, angle specifies the bending angle, and xyz determines the center position of the structure in three directions. z_span defines the thickness of the waveguide.
st.add_geometry(name="ring", type="Ring", property={
"material": {"material": mt["Si"], "mesh_order": 2},
"geometry": {"x": 1, "y": 2*space,"z": 1, "z_span": wg_height,
"angle":70,
"inner_radius": size-wg_width/2, "outer_radius": size+wg_width/2,
"tilt_angle1": 70, "tilt_position": "bottom", "tilt_angle2": 90,}})
3.4 How to add a circular structure?
Next, we'll learn how to add a circular structure using the following code. In this code, radius represents the radius of the circle, and x/y denote the coordinates of the center of the circle.
st.add_geometry(name="circle", type="Circle", property={
"material": {"material": mt["Si"], "mesh_order": 2},
"geometry": {"radius": size,"angle":90,
"x": 2*space, "y": 1,"z": 1,"z_span": wg_height,
"tilt_angle":70,"tilt_position":"middle",}})
3.6 How to add a linear taper (Taper) structure?
To create a linear taper structure, use the add_geometry function with type="LinearTrapezoid". Specify the coordinates of the four vertices of the taper structure using control_points. x/y/z determine the 3D reference point of the taper structure, and z_span sets the thickness of the taper waveguide.
st.add_geometry(name="linear_trapezoid", type="LinearTrapezoid", property={
"material": {"material": mt["Si"], "mesh_order": 2},
"geometry": {"x": 2*space, "y": space,"z": 1, "z_span": wg_height,
"control_points":[{"x": 1, "y": 1}, {"x": 1, "y": 2}, {"x": 2, "y": 2}, {"x": 2, "y": 0.1}],
"tilt_angle": 70, "tilt_position": "bottom",}})
3.7 How to add a custom function-based geometric structure?
Let's take the example of a taper structure with a custom function curve. To do this, use the add_geometry function with type="AnalyticalWaveguide". In this case, equation1 represents the expression of the waveguide boundary function. The parameters x/y/z denote the center coordinates, x span/y span define the domain range, resolution sets the resolution, nonsymmetric determines whether the structure is non-symmetric.
st.add_geometry(name="analytical_waveguide",type="AnalyticalWaveguide",
property={
"material": {"material": mt["Si"], "mesh_order": 2},
"geometry": {
"equation1":"x^0.5",
"nonsymmetric": True,
"equation2":"x",
"x": 5*space, "x_span":20,
"y":5 *space, "y_span":20,
"z": 0, "z_span": wg_height,
"resolution":20,
"tilt_angle":70,"tilt_position":"middle",}})
3.8 How to import a GDS file and create the corresponding model in SDK?
The following code example demonstrates how to import a GDS file and create the corresponding structure in SDK. First, we need to obtain the full file path of the GDS file (gds_file). Then, use the add_geometry function with type="gds_file" to import the GDS file. The path parameter should contain the full path to the GDS file, cell_name is the name of the cell to be imported from the GDS file, layer_name specifies the layer type and data type to be imported, and material is the material associated with the structure.
gds_file_root_path = os.path.abspath(os.path.join(path, '..'))
gds_file = gds_file_root_path + "/examples_gds/fast_fdtd.gds"
st.add_geometry(name="gds_file_3D",type="gds_file",
property={
"material": {"material": mt["Si"], "mesh_order": 2},
"general": {"path": gds_file, "cell_name": "EXTEND_1", "layer_name": (3, 0)},
"geometry": {"x": 4*space, "y": 2*space,"z": 0.05,"z_span": 0.1,
"tilt_angle": 60,"tilt_position": "bottom",}})
During the process of modeling by importing GDS files, it's important to pay attention to the setting of the material's mesh_order to ensure that the overlapping structures are covered in the correct order.
3.9 How to set the mesh order?
The mesh order decides the coverage when creating a geometric structure.
When the mesh order of two structures are same, the structure which is established later has a higher priority. When the mesh order of two structures are different, the large numerical value of mesh order has greater priority than the small one. That is, The large mesh order of structure is able to cover small mesh order of structure. For example, the mesh order=2 structure will cover the mesh order=1.
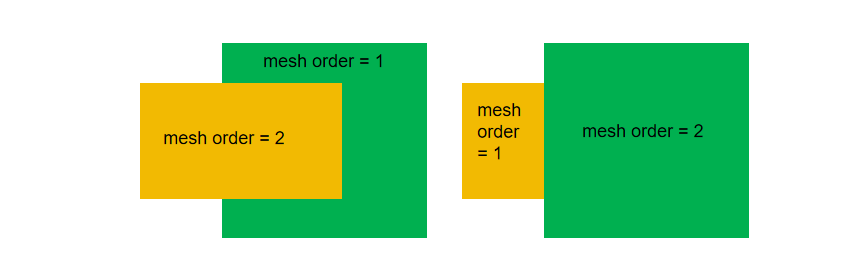
The advantage is that increasing the value of mesh order allows user to make new nested structures in the complex model.
4. How to set up FDE simulation in SDK?
4.1 How to configure various parameters for FDE simulation?
Next, we will learn how to add an FDE simulation and set its simulation parameters in SDK using the Simulation function.
Within the simu.add section, we can set the simulation name name, simulation type type, and various simulation parameters in the property field. Within property. And we also need to set the simulation boundary, where geometry defines the geometry parameters, and boundary_conditions specifies the cross-sectional simulation boundary parameters, we can set the simulation solver type through solver_type (with the default value as 2d_x_normal). In the mesh_settings section, we can configure mesh parameters, including setting the grid sizes in different directions using global_mesh_uniform_grid.
# region --- Simulation ---
simu = pj.Simulation()
simu.add(name=simu_name, type="FDE",
property={"background_material": mt["Air"],
"geometry": {"x": 0, "x_span": 0, "y": 0, "y_span": yspan_solver, "z": 0, "z_span": zspan_solver, },
"boundary_conditions": {"y_min_bc": "PML", "y_max_bc": "PML", "z_min_bc": "PML", "z_max_bc": "PML",
"pml_settings": {"pml_layer": 12, "pml_kappa": 2, "pml_sigma": 5,
# "polynomial_order": 3,
}},
# 'mode_removal': {'threshold': 0.02},
'fractional_offset_for_group_delay': 0.0003,
'general': {'solver_type': '2d_x_normal'}, # default is '2d_x_normal' ['2d_x_normal','2d_y_normal','2d_z_normal']
"mesh_settings": {"mesh_refinement": {"mesh_refinement": "curve_mesh"}, "mesh_factor": 1.2,
"global_mesh_uniform_grid": {"dy": grid, "dz": grid, },
# 'minimum_mesh_step_settings': {'min_mesh_step': 1.0e-4}
}})
simu_res = simu[simu_name].run()
# endregion
4.2 How to calculate the overlap in FDE simulation?
Overlap calculations in FDE simulation can be performed by utilizing the following code to calculate the overlap between the mode light and a Gaussian light source, as illustrated in the code below.
if run_options.run_overlap:
if run_options.run_beam:
beam_res = simu[simu_name].run_fde_beam_and_extract(
property={"define_gaussian_beam_by": "waist_size_and_position", # [waist_size_and_position,beam_size_and_divergence],
"waist_radius": 5.2, "distance_from_waist": 1.5, "refractive_index": 1.45, "theta": 0, "phi": 0,
"polarization_angle": 90, "sample_span": 6, "sample_resolution": 200},
savepath=plot_path + "beam_heatmap")
analysis.add(name="overlap", type="overlap",
property={"field_1": {"workflow_id": beam_res.workflow_id, "mode": 0},
"field_2": {"workflow_id": fde_res.workflow_id, "mode": 0},
"optimize_position": True})
overlap_res = analysis["overlap"].run()
overlap_res.extract(
export_csv=True, savepath=plot_path + "overlap")
4.3 How to view the refractive index profile in the FDE/FDTD/EME modules?
As demonstrated in the code below, we can visualize the refractive index profile of a device's cross-section using index monitor in FDTD/EME simulation.Assuming that we want to calculate the refractive index profile on the x-direction cross-section of the device. Set the position of the index monitor along the x-axis by specifying x=0 and setting x_span=0. Then, configure the center coordinates y/z and the dimensions y_span/z_span for the cross-sectional refractive index profile calculation area. The setup for calculating the refractive index profile in the y/z-direction cross-section is analogous.
mn = pj.Monitor()
mn.add(name="index_monitor_1",type="index_monitor",
property={"geometry": { "x": 0, "x_span": 0, "y": 0, "y_span": 5, "z": 0, "z_span": 5,}},)
# results view
simu[simu_name].preview_index(monitor_name="x_normal_index", savepath=f"{plot_path}01_IndexPreview_x=0")
And in the FDE simulation, we can use the mesh structure to preview the index directly.
analysis = pj.Analysis()
analysis.add_analysis(name="fast_FDE_Analysis", type="FDEAnalysis",
props={"workflow_id": simu_res.workflow_id, "simulation_name": "FDE",
"modal_analysis": {"calculate_modes": run_options.run, "mesh_structure": True,
"wavelength": wavelength, "number_of_trial_modes": number_of_trial_modes,
"search": "max_index", # ['near_n','max_index']
# "n": 1,
"calculate_group_index": False,
"bent_waveguide": {"bent_waveguide": False, "radius": 1, "orientation": 0, "location": "simulation_center"}
},
"frequency_analysis": {"frequency_analysis": run_options.run_frequency_sweep,
"start_wavelength": 1.50, "stop_wavelength": 1.60, "number_of_points": 3,
"effective_index": 1, "detailed_dispersion_calculation": False
}})
result_fde = analysis["fast_FDE_Analysis"].run()
4.4 How to retrieve simulation data in the FDE module of SDK?
To obtain simulation data in the FDE module of SDK, we can first use result_fde.extract with data='calculate_modes' to retrieve neffctive table of modes, which can be saved in the savepath directory. we can choose to export the mode field data as a CSV file by using export_csv. Set the attribute, mode, real, and imag parameters to extract the specific mode-related data we need. When using the frequency scan feature in the FDE module, we can extract data by specifying data="frequency_analysis". As shown in the code, we can retrieve various components such as "neff," "loss," "group_index," "polarization," and more.
# region --- See Results ---
if run_options.extract:
if run_options.run:
result_fde.extract(
data="mesh_structure", savepath=f"{plot_path}01_index", export_csv=True)
res = result_fde.extract(
data="calculate_modes", savepath=f"{plot_path}02_neff_table", export_csv=True)
print(res.to_string(index=True))
for m in range(len(res)):
result_fde.extract(data="calculate_modes", savepath=f"{plot_path}03_mode{m}",
attribute="E", mode=m, real=True, imag=True, **export_options, show=False)
if run_options.run_frequency_sweep:
attr_selections: List[Literal["neff", "loss", "group_index", "polarization"]] = [
"neff", "loss", "group_index", "polarization"]
for i, a in enumerate(attr_selections):
result_fde.extract(data="frequency_analysis", savepath=f"{plot_path}04_freq_sweep_{a}",
attribute=a, real=True, imag=True, export_csv=True, export_mat=True, show=False)
# endregion
5. How to set up EME simulation in SDK?
5.1 How to configure boundary conditions and ports in EME simulation?
To begin with, we need to define the boundary conditions. Then, we can set the geometric dimensions of the boundaries with geometry and configure the boundary parameters for each dimension with boundary_conditions in the EME simulation.
Next, we should configure the EME ports using Port. we can add a new port using add. Specify the port_location for the port's location. we can choose the mode for this port through mode_selection, such as fundamental_TE or fundamental_TM.
Additionally, we can select user_select to define a custom mode for the port by setting mode_index to a positive integer value. Use use_full_simulation_span to determine whether the full simulation span should be used.
# region --- Simulation ---
simu = pj.Simulation()
simu.add(name=simu_name, type="EME",
property={"background_material": mt["SiO2"],
"mesh_settings": {"mesh_factor": 1.2, "mesh_refinement": {"mesh_refinement": "curve_mesh"}},
"geometry": {"x_min": -1, "y": 0, "y_span": 3, "z": 0, "z_span": 3},
"boundary_conditions": {"y_min_bc": "PML", "y_max_bc": "PML", "z_min_bc": "PML", "z_max_bc": "PML",
"pml_settings": {"pml_kappa": 2, "pml_sigma": 5, "pml_layer": 12, "pml_polynomial": 3}
},
"general": {"wavelength": wavelength, "wavelength_offset": 0.0003, "use_wavelength_sweep": True},
"eme_setup": {"cell_geometry": {"allow_custom_eigensolver_settings": True,
"cell_group_definition": [{"span": 2, "cell_number": cell_number, "number_of_modes": number_of_modes, "sc": "sub_cell"}]}},
"transverse_mesh_setting": {"global_mesh_uniform_grid": {"dy": grid, "dz": grid}}
})
# endregion
# region --- EME Port ---
pjp = pj.Port()
pjp.add(name="eme_in", type="eme_port",
property={"modal_analysis": {"wavelength": wavelength},
"geometry": {"port_location": "left",
# 'use_full_simulation_span': False, # default is 'True' # "y": 0, # "y_span": 3, # "z": 0, # "z_span": 3
},
"eme_port": {"general": {"mode_selection": ("fundamental_TE"), "number_of_trial_modes": number_of_modes,
# 'mode_index': 0, 'search': 'max_index'
}}})
pjp.add(name="eme_out", type="eme_port",
property={"modal_analysis": {"wavelength": wavelength, },
"geometry": {"port_location": "right",
# 'use_full_simulation_span': False, # default is 'True'
# "y": 0,"y_span": 3,"z": 0,"z_span": 3
},
"eme_port": {"general": {"mode_selection": ("fundamental_TE"), "number_of_trial_modes": number_of_modes,
# 'mode_index': 0, 'search': 'max_index'
}}})
# endregion
5.2 How to add a profile monitor in EME simulation?
As shown in the code below, we can add a new monitor to wer EME simulation. Use type='profile_monitor' to specify the type of monitor we want to add. In the property section, we can configure various parameters for the monitor. For example, set monitor_type to determine the direction of the monitor. Define the coordinates and dimensions of the monitor using x/y/z and x_span/y_span/z_span, with the normal span width set to zero.
# region --- Monitor ---
mn = pj.Monitor()
mn.add(name="x_normal", type="profile_monitor",
property={"geometry": {"monitor_type": "2d_x_normal",
# 'x_resolution': 100,
"x": 0.9, "x_span": 0, "y": 0, "y_span": 3, "z": 0, "z_span": 3}})
# endregion
5.3 How to configure parameters for EME simulation?
Next, we will learn how to set the parameters for EME simulation within the code below. Under the general parameters, we can define the wavelength for the EME simulation wavelength.
Following that, we can define the EME simulation cells using cell_group_definition. Within this section, span represents the length of each cell, cell_number specifies how many cells the length should be evenly divided into, number_of_modes determines the number of modes to be solved during the calculation, and sc determines the method for calculating the S-matrix between cells.
The transverse_mesh_setting section allows we to configure the transverse mesh settings.
# region --- Simulation ---
simu = pj.Simulation()
simu.add(name=simu_name, type="EME",
property={"background_material": mt["SiO2"],
"mesh_settings": {"mesh_factor": 1.2, "mesh_refinement": {"mesh_refinement": "curve_mesh"}},
"geometry": {"x_min": -1, "y": 0, "y_span": 3, "z": 0, "z_span": 3},
"boundary_conditions": {"y_min_bc": "PML", "y_max_bc": "PML", "z_min_bc": "PML", "z_max_bc": "PML",
"pml_settings": {"pml_kappa": 2, "pml_sigma": 5, "pml_layer": 12, "pml_polynomial": 3}
},
"general": {"wavelength": wavelength, "wavelength_offset": 0.0003, "use_wavelength_sweep": True},
"eme_setup": {"cell_geometry": {"allow_custom_eigensolver_settings": True,
"cell_group_definition": [{"span": 2, "cell_number": cell_number, "number_of_modes": number_of_modes, "sc": "sub_cell"}]}},
"transverse_mesh_setting": {"global_mesh_uniform_grid": {"dy": grid, "dz": grid}}
})
# endregion
5.4 How to set up a preview of EME simulation structure images?
we can preview the EME simulation structure and images using structure_show. In the example code below, the parameter fig_type determines the image format, typically "png." Use show to specify whether we want to generate images. Set savepath for the path where the result images will be saved. we can use celldisplay to control whether cells are displayed in the image, and xyratio adjusts the aspect ratio between the x and y coordinates in the image, with the default value being (1,1).
# region --- Structure Show ---
st.structure_show(fig_type="png", show=False,
savepath=f"{plot_path}00_{simu_name}", celldisplay=True, xyratio=(1, 1))
# endregion
5.5 How to obtain EME port mode results?
we can retrieve the results of EME prot mode by using preview_modes. In the example code below, we can extract the port mode information. Set data="calculate_modes" to specify that we want to retrieve EME port mode results. Use save_path to specify the data saving path, and define attribute to specify the data content we wish to extract.
# region --- 11. Calculate Mode ---
if run_options.calculate_modes:
for port in ["eme_in", "eme_out"]:
simu[simu_name].preview_modes(port_name=port, data="calculate_modes",
savepath=f"{plot_path}02_modeprofile_fdeonly_{port}", attribute="E", mode=0)
simu[simu_name].preview_modes(port_name=port, data="calculate_modes",
savepath=f"{plot_path}02_Preview_{port}_neff", show=False, export_csv=True)
# endregion
5.6 How to obtain mode field plots for EME/FDTD?
Similarly, we can use eme_res.extract to retrieve simulation results. In the example code below, we can set data='eme_propagate:monitor' to specify that we want to obtain monitor results. Define the save_path for data storage and use the attribute parameter to specify the data to be extracted. plot_x and plot_y control the x and y-axis coordinates for line or heatmap plots. Use real and imag to determine how the output data should be handled: when both are False, the program will raise an error with "real and imag are both false"; when both are True, the results will be taken as the absolute value; if one is True, either the real or imaginary part will be output. show controls whether images are displayed, and export_csv determines if data should be saved as a CSV file.
eme_res.extract(data="eme_propagate:monitor", savepath=plot_path + "013_eme_z_normal",
monitor_name="z_normal", attribute="E",
# plot_x='y', plot_y='z',
# real=True, imag=True,
# export_csv=False, show=False
)
5.7 How to perform EME length sweeps?
As demonstrated in the code below, we can generate N x N S-parameter matrix length sweep plots using data="propagation_sweep:sweep", where N represents the number of ports.
# region --- EME Propagation Sweep Results ---
if run_options.run_length_sweep:
eme_res.extract(
data="propagation_sweep:sweep",
savepath=f"{plot_path}{kL[4]}_length_sweep",
export_csv=True,
)
# endregion
5.8 How to perform EME wavelength sweeps?
As shown in the code below, we can generate N x N S-parameter matrix wavelength sweep plots using data="wavelength_sweep:sweep", where N represents the number of ports.
# region --- EME Wavelength Sweep Results ---
if run_options.run_wavelength_sweep:
eme_res.extract(data="wavelength_sweep:sweep", savepath=plot_path +
"06_wavelength_sweep", plot_x="wavelength", export_csv=True)
# endregion
5.9 During EME simulations, is overlap recalculated when wavelength scans are repeated?
During EME simulations, when performing wavelength sweeps, the workflow involves first computing all the modes within the cells, and then during the sweep, calculating overlap and normalization as needed. If we repeat wavelength scans, there is no need to recalculate overlap. The program will perform the overlap calculation only once.
6. How to configure FDTD simulations in SDK?
6.1 How to set the wavelength for FDTD simulations?
Waveform supports defining wavelength, wavelength span, and related parameters. It includes common communication wavelengths like 1550 nm and 1310 nm, as well as visible light wavelengths.
# region --- Waveform ---
wv = pj.Waveform()
wv.add(name=waveform_name, type='gaussian_waveform',
property={'set': 'frequency_wavelength', # selections are ['frequency_wavelength','time_domain']
'set_frequency_wavelength': {
'range_type': 'wavelength', # selections are ['frequency','wavelength']
'range_limit': 'center_span', # selections are ['min_max','center_span']
'wavelength_center': wavelength,
'wavelength_span': 0.1,},})
# endregion
6.2 How to set up light sources for FDTD simulations?
we can configure light sources for FDTD simulations using the source function, as shown in the code below. In this setup, the type specifies the source type, commonly using mode sources in waveguides and Gaussian sources in free space. The inject_axis and direction determine the reference propagation dierection for the light source. we can select specific modes using mode_selection and mode_index, and choose a specific waveform with waveform. The geometry parameter defines the geometric dimensions of the light source.
# region --- ModeSource ---
src = pj.Source()
src.add(name="source", type="mode_source",
property={"general": {"mode_selection": "fundamental_TE", "waveform": {"waveform_id": wv[waveform_name]}, "inject_axis": "x_axis", "direction": "forward"},
"geometry": {"x": -l_input-l_beam/2-l_bend+2, "x_span": 0, "y": 1.35, "y_span": monitor_w, "z": 0.11, "z_span": monitor_h}})
# endregion
6.3 How to configure monitors in FDTD simulations?
Let's briefly introduce the setup of FDTD monitors, including global monitors, power monitors, and mode expansion monitors. As shown in the code below, we can use type='global_option' to select a global monitor. Set wavelength_center to specify the center wavelength for the monitor, wavelength_span for the wavelength range, and frequency_points to determine the number of points monitored within that wavelength range.
# region --- GlobalMonitor ---
mn = pj.Monitor()
mn.add(name="Global Option", type="global_option",
property={"frequency_power": {"spacing_type": "wavelength", "spacing_limit": "center_span",
"wavelength_center": wavelength, "wavelength_span": 0.1, "frequency_points": 11}})
# endregion
As shown in the following code, we can set up power monitors in FDTD simulations using the type='power_monitor'. The name parameter specifies the monitor's name. In the property section, we can configure various monitor parameters, including wavelength-related settings such as wavelength_center, wavelength_span, and frequency_points. The geometry section allows we to set the monitor's orientation, position, and dimensions with parameters such as monitor_type, x/y/z, and x_span/y_span/z_span.
# region --- Through ---
mn.add(name="Znormal", type="power_monitor",
property={"general": {"frequency_profile": {"wavelength_center": wavelength, "wavelength_span": 0.1, "frequency_points": 3}},
"geometry": {"monitor_type": "2d_z_normal", "x_min": -(l_input+l_bend+l_beam/2-0.5), "x_max": l_input+l_bend+l_beam/2-0.5, "y": 0, "y_span": 5, "z": 0.11, "z_span": 0}})
# endregion
6.4 How to configure Port-related parameters in FDTD?
we can set the parameters related to ports in FDTD simulations as demonstrated in the code below. The waveform_id specifies the input wavelength, and source_port determines the input port. Then, using the add method, we can add the corresponding port with type='fdtd_port', providing a name for the port. In the property section, we can configure its relevant parameters, and in the geometry section, we can set the port's position and dimensions. The modal_properties parameter allows we to specify the properties of the input light, including inject_axis for the light's injection axis, direction for the direction of the incident light, and mode_selection for selecting the light mode.
# region --- Port ---
pt = pj.Port(property={"waveform_id": wv[waveform_name],
"source_port": "left_port", "monitor_frequency_points": 11})
pt.add(name="left_port", type="fdtd_port",
property={"geometry": {"x": -wg_length / 2 + span, "x_span": 0, "y": 0, "y_span": port_width, "z": 0, "z_span": port_height, },
"modal_properties": {"general": {"inject_axis": "x_axis", "direction": "forward", "mode_selection": "fundamental"}}})
# endregion
6.5 How to extract relevant results from FDTD simulations?
As shown in the following code, we can specify the savepath for saving the results. Use target='line' for line plots or target='intensity' for intensity plots. The attribute parameter specifies the parameters to be extracted, and wavelength is used to specify the wavelength.
To extract the mode field at a specific wavelength from a power monitor:
fdtd_res.extract(data="fdtd:power_monitor", savepath=f"{plot_path}04_x_normal_abs(E)",
target="intensity", attribute="E", monitor_name="x_normal", wavelength=str(wavelength), export_csv=True)
Extracting the transmittance at different wavelengths from a power monitor:
fdtd_res.extract(data="fdtd:power_monitor", savepath=f"{plot_path}03_x_normal_abs(T)",
target="line", attribute="T", monitor_name="x_normal", plot_x="wavelength", export_csv=True)
Extracting the response of a mode expansion monitor to a specific mode transmission from a power monitor:
me_res.extract(data="fdtd:mode_expansion", savepath=f"{plot_path}04_TransVsOrder",
target="line", attribute="T_forward", real=True, imag=True, mode_expansion_name="me_through", wavelength=f"{wavelength}", plot_x="mode", show=False, export_csv=True)
Extracting the S-matrix results from FDTD simulations:
smatrix_res.extract(data="smatrix_sweep", savepath=f"{plot_path}05_smatrix_sweep",
target="line", plot_x="wavelength", export_csv=True)
6.6 Why does the S-matrix in FDTD simulation sometimes exceed 1?
In FDTD simulations, the presence of a light source in the simulation region can lead to incorrect S-matrix results, such as some port responses exceeding 1. To ensure accurate S-matrix calculations, it's essential to make sure that there are no light sources within the simulation region.